Please click here to go to the tutorial for DIALS 2.2.
Refining multi-tile detector metrology with DIALS¶
Warning
This tutorial is out of date! Default parameters and output have changed.
Introduction¶
At the end of the Processing in Detail, we showed plots from dials.analyse_output, including images of the positional residuals as a function of position on the detector face. There were clearly some systematic effects, suggesting whole-tile shifts or rotations. Generally, these are small (typically much less than the pixel size) and are expected. DECTRIS provides calibration tables from factory metrology experiments, which XDS can use. Currently we are not applying the correction tables to reflection positions in DIALS. In some situations we may have to work with a much less well calibrated or characterised detector than the Pilatus instruments installed at Diamond. Nevertheless, the multi-panel detector models in DIALS allow us to discover the appropriate corrections assuming we have good diffraction data to refine against.
Here we have access to a thaumatin dataset collected with low dose to avoid radiation damage and with the detector 400 mm from the sample to ensure the coverage of reflections extends right out towards the corners of the images.
Preparing for multi-tile refinement¶
Usually multi-panel detectors like the Pilatus P6M are treated as if they were single panels (with dead regions) in DIALS. Clearly, if we are to make corrections to the relative positions and orientations of the panels, we need a more detailed detector model in which the panels are treated separately. Currently there isn’t really a neat way to switch between the single and multiple panel models, but until we have a native mechanism for doing this in DIALS there is a hidden workaround that does this. We just need to set an environment variable:
export P6M_60_PANEL=1
With that in place, we start as usual by importing the dataset:
dials.import Thaum_M10S15_3_*.cbf
The fact that the multiple panel detector model was used is clear from the output of:
dials.show imported.expt
We can now inspect images from the dataset with:
dials.image_viewer imported.expt
However, the dataset is at such low dose that it can be difficult to find the spots. It might be helpful to us a more mature image viewer such as ADXV or Albula for this.
We have to run spot finding throughout the dataset. It is a 360 degree sequence so this will take a few minutes. We can use more processes to move a little quicker:
dials.find_spots imported.expt nproc=8 min_spot_size=3
Here we chose to set min_spot_size=3, which overrides the default of 6 used
for this detector model. We did this because otherwise this weakly diffracting
dataset produces rather few strong spots, whereas we want as much coverage as
we can get, right out to the corners
Now to index the data. Although it is a well-diffracting crystal, running indexing with defaults finds an approximate supercell with the a and b lengths slightly more than doubled. Inspection of the found spots with dials.reciprocal_lattice_viewer shows curved lines of spots rather than a regular grid, indicating a poor starting model. There are plenty of things that can be done to get indexing to find the correct cell here. If you have time you might like to try a few options out. However, if not, we won’t worry too much about it, because if we pass in the known unit cell for thaumatin then it works just fine. We also chose to apply tetragonal symmetry immediately:
dials.index imported.expt strong.refl space_group="P 4" unit_cell="58 58 150 90 90 90"
The output of refinement in the highest resolution macrocycle is as follows:
################################################################################
Starting refinement (macro-cycle 5)
################################################################################
Summary statistics for observations matched to predictions:
--------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
--------------------------------------------------------------------------
| Xc - Xo (mm) | -2.492 | -0.04261 | -0.0004832 | 0.0424 | 2.131 |
| Yc - Yo (mm) | -1.697 | -0.03481 | 0.001733 | 0.03682 | 1.656 |
| Phic - Phio (deg) | -0.9645 | -0.06454 | -0.0007655 | 0.06379 | 0.8353 |
| X weights | 80.55 | 127.8 | 131.8 | 133.9 | 135.2 |
| Y weights | 77.02 | 127.6 | 131.8 | 134 | 135.2 |
| Phi weights | 26.58 | 32.59 | 32.65 | 32.65 | 32.65 |
--------------------------------------------------------------------------
Refinement steps:
-------------------------------------------------
| Step | Nref | RMSD_X | RMSD_Y | RMSD_Phi |
| | | (mm) | (mm) | (deg) |
-------------------------------------------------
| 0 | 18025 | 0.067273 | 0.063827 | 0.086195 |
| 1 | 18025 | 0.067369 | 0.063706 | 0.086197 |
| 2 | 18025 | 0.067439 | 0.063629 | 0.086155 |
| 3 | 18025 | 0.067531 | 0.063529 | 0.086115 |
| 4 | 18025 | 0.067587 | 0.063456 | 0.086122 |
| 5 | 18025 | 0.067607 | 0.063419 | 0.086155 |
| 6 | 18025 | 0.067617 | 0.063401 | 0.086176 |
| 7 | 18025 | 0.06762 | 0.063395 | 0.086181 |
-------------------------------------------------
RMSD no longer decreasing
RMSDs by experiment:
----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
----------------------------------------------
| 0 | 18025 | 0.39314 | 0.36858 | 0.24623 |
----------------------------------------------
RMSDs by panel:
-----------------------------------------------
| Panel | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
-----------------------------------------------
| 0 | 10 | 0.53322 | 0.82846 | 0.14379 |
| 1 | 140 | 0.37822 | 0.43153 | 0.26887 |
| 2 | 297 | 0.41585 | 0.40848 | 0.26118 |
| 3 | 146 | 0.5563 | 0.40867 | 0.27493 |
| 4 | 17 | 0.49115 | 0.43483 | 0.21656 |
| 5 | 44 | 0.41713 | 0.46071 | 0.27804 |
| 6 | 405 | 0.44932 | 0.51832 | 0.26569 |
| 7 | 758 | 0.33153 | 0.28766 | 0.25461 |
| 8 | 478 | 0.38555 | 0.40525 | 0.26244 |
| 9 | 96 | 0.47338 | 0.45054 | 0.24652 |
| 10 | 152 | 0.40191 | 0.959 | 0.2976 |
| 11 | 701 | 0.31004 | 0.3035 | 0.24597 |
| 12 | 1128 | 0.37366 | 0.24303 | 0.24016 |
| 13 | 802 | 0.33404 | 0.32094 | 0.24614 |
| 14 | 201 | 0.50746 | 0.47124 | 0.27169 |
| 15 | 231 | 0.40185 | 0.56122 | 0.3029 |
| 16 | 745 | 0.21926 | 0.30075 | 0.21426 |
| 17 | 831 | 0.17864 | 0.17706 | 0.2176 |
| 18 | 778 | 0.36795 | 0.21167 | 0.20696 |
| 19 | 269 | 0.44757 | 0.40028 | 0.23809 |
| 20 | 205 | 0.48078 | 0.65283 | 0.34241 |
| 21 | 467 | 0.37577 | 0.35789 | 0.20992 |
| 22 | 370 | 0.22329 | 0.29697 | 0.20943 |
| 23 | 447 | 0.36708 | 0.22722 | 0.20176 |
| 24 | 231 | 0.28466 | 0.52795 | 0.31729 |
| 25 | 3 | 0.36377 | 1.1547 | 0.70452 |
| 26 | 28 | 0.14842 | 0.29869 | 0.23437 |
| 27 | 22 | 0.13375 | 0.2666 | 0.17175 |
| 28 | 17 | 0.1222 | 0.14108 | 0.17261 |
| 29 | 4 | 0.40725 | 0.85708 | 0.42044 |
| 30 | 145 | 0.60805 | 0.57195 | 0.43388 |
| 31 | 281 | 0.18353 | 0.21351 | 0.2061 |
| 32 | 250 | 0.19984 | 0.15417 | 0.1993 |
| 33 | 294 | 0.15006 | 0.27665 | 0.21823 |
| 34 | 158 | 0.44084 | 0.50436 | 0.38365 |
| 35 | 203 | 0.86526 | 0.47248 | 0.32568 |
| 36 | 525 | 0.54992 | 0.31614 | 0.19036 |
| 37 | 521 | 0.22611 | 0.12494 | 0.21894 |
| 38 | 624 | 0.23105 | 0.20793 | 0.22246 |
| 39 | 259 | 0.43408 | 0.43312 | 0.2724 |
| 40 | 134 | 0.55289 | 0.3935 | 0.26933 |
| 41 | 557 | 0.29543 | 0.24331 | 0.23127 |
| 42 | 771 | 0.46252 | 0.19655 | 0.2348 |
| 43 | 658 | 0.24901 | 0.31232 | 0.24217 |
| 44 | 199 | 0.44994 | 0.43547 | 0.28695 |
| 45 | 54 | 0.5679 | 0.44719 | 0.23983 |
| 46 | 333 | 0.44117 | 0.44019 | 0.24586 |
| 47 | 592 | 0.3056 | 0.44653 | 0.24795 |
| 48 | 411 | 0.67885 | 0.39219 | 0.2588 |
| 49 | 93 | 0.45355 | 0.5659 | 0.2611 |
| 50 | 5 | 1.0009 | 0.46232 | 0.21423 |
| 51 | 162 | 0.72162 | 0.36619 | 0.23884 |
| 52 | 324 | 0.51574 | 0.48569 | 0.2488 |
| 53 | 224 | 0.41145 | 0.40151 | 0.2671 |
| 54 | 16 | 0.5362 | 0.50568 | 0.21454 |
| 55 | 1 | 0.85351 | 0.57076 | 0.24389 |
| 56 | 37 | 0.72428 | 0.38373 | 0.27121 |
| 57 | 106 | 0.80208 | 0.98469 | 0.27939 |
| 58 | 61 | 0.5144 | 1.3281 | 0.30346 |
| 59 | 4 | 0.33371 | 0.50058 | 0.24425 |
-----------------------------------------------
Final refined crystal models:
model 1 (192715 reflections):
Crystal:
Unit cell: (57.834, 57.834, 150.022, 90.000, 90.000, 90.000)
Space group: P 4
U matrix: {{ 0.4122, -0.9018, 0.1299},
{ 0.2361, -0.0320, -0.9712},
{ 0.8800, 0.4310, 0.1997}}
B matrix: {{ 0.0173, 0.0000, 0.0000},
{ 0.0000, 0.0173, 0.0000},
{ 0.0000, 0.0000, 0.0067}}
A = UB: {{ 0.0071, -0.0156, 0.0009},
{ 0.0041, -0.0006, -0.0065},
{ 0.0152, 0.0075, 0.0013}}
This refinement was performed moving all the panels as a rigid block, as usual.
With overall positional RMSDs within 40% of the pixel size and a
quarter of the image width in \(\phi\) we can see straight away that we are
dealing with a fairly good quality
dataset. There are a few outliers of well over 1 mm on the detector surface and nearly
1 degree in \(\phi\) though, which we would prefer not to include in
refinement. The outliers are not as bad if we had kept min_spot_size=6,
but the detector coverage is worse in that case. Although from the indexing results
it seems that coverage of reflections on the outer panels is rather low, so far
we let refinement take a random subset of the data in order to index quicker,
so there’s no need to worry about that yet.
Now we will refine the detector as a rigid block again, turning on outlier
rejection and requesting to use all reflections to get the best we can out
of the dataset. We will also keep the refined reflections file for analysis.
The final parameter here, close_to_spindle_cutoff=0.01 allows reflections
closer to the spindle to be included in refinement (default value is 0.05, and
if set to 0.0 no reflections will be rejected for being too close).
Without this option, the central panels are very sparse:
dials.refine indexed.refl indexed.expt \
outlier.algorithm=tukey use_all_reflections=true close_to_spindle_cutoff=0.01 \
output.reflections=refined_lev0.refl \
output.experiments=refined_lev0.expt
Here is the output:
The following parameters have been modified:
output {
reflections = refined_lev0.refl
experiments = refined_lev0.expt
}
refinement {
reflections {
use_all_reflections = true
close_to_spindle_cutoff = 0.01
outlier {
algorithm = null *tukey
}
}
}
input {
experiments = indexed.expt
reflections = indexed.refl
}
Configuring refiner
Summary statistics for observations matched to predictions:
-----------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
-----------------------------------------------------------------------
| Xc - Xo (mm) | -2.487 | -0.04174 | 0.000276 | 0.0424 | 2.128 |
| Yc - Yo (mm) | -1.75 | -0.03468 | 0.00168 | 0.03656 | 1.654 |
| Phic - Phio (deg) | -5.549 | -0.06627 | -0.002108 | 0.063 | 3.27 |
| X weights | 80.55 | 127.9 | 131.9 | 134 | 135.2 |
| Y weights | 77.02 | 127.7 | 131.9 | 134 | 135.2 |
| Phi weights | 25.97 | 32.57 | 32.65 | 32.65 | 32.65 |
-----------------------------------------------------------------------
6375 reflections have been rejected as outliers
Summary statistics for observations matched to predictions:
--------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
--------------------------------------------------------------------------
| Xc - Xo (mm) | -0.2933 | -0.04128 | -3.167e-05 | 0.04112 | 0.3725 |
| Yc - Yo (mm) | -0.4707 | -0.03376 | 0.001821 | 0.03588 | 0.4945 |
| Phic - Phio (deg) | -0.6965 | -0.06574 | -0.002213 | 0.06229 | 0.7919 |
| X weights | 80.55 | 128 | 132 | 134 | 135.2 |
| Y weights | 77.02 | 127.8 | 131.9 | 134.1 | 135.2 |
| Phi weights | 26.58 | 32.57 | 32.65 | 32.65 | 32.65 |
--------------------------------------------------------------------------
Performing refinement...
Refinement steps:
--------------------------------------------------
| Step | Nref | RMSD_X | RMSD_Y | RMSD_Phi |
| | | (mm) | (mm) | (deg) |
--------------------------------------------------
| 0 | 186203 | 0.064091 | 0.057786 | 0.086136 |
| 1 | 186203 | 0.064045 | 0.057829 | 0.08608 |
| 2 | 186203 | 0.063949 | 0.05791 | 0.086068 |
| 3 | 186203 | 0.063825 | 0.058023 | 0.086026 |
| 4 | 186203 | 0.063734 | 0.058114 | 0.085958 |
| 5 | 186203 | 0.063682 | 0.058167 | 0.085909 |
| 6 | 186203 | 0.063654 | 0.058198 | 0.085887 |
| 7 | 186203 | 0.063645 | 0.058208 | 0.085882 |
| 8 | 186203 | 0.063644 | 0.05821 | 0.085882 |
--------------------------------------------------
RMSD no longer decreasing
RMSDs by experiment:
-----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
-----------------------------------------------
| 0 | 186203 | 0.37002 | 0.33843 | 0.24538 |
-----------------------------------------------
RMSDs by panel:
-----------------------------------------------
| Panel | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
-----------------------------------------------
| 0 | 63 | 0.47216 | 0.59331 | 0.22352 |
| 1 | 1363 | 0.35019 | 0.46301 | 0.26549 |
| 2 | 3076 | 0.4128 | 0.40658 | 0.26659 |
| 3 | 1630 | 0.46232 | 0.39692 | 0.25547 |
| 4 | 112 | 0.39711 | 0.42575 | 0.23841 |
| 5 | 394 | 0.35145 | 0.51263 | 0.25474 |
| 6 | 3489 | 0.3238 | 0.48667 | 0.25694 |
| 7 | 6172 | 0.30869 | 0.27402 | 0.25323 |
| 8 | 3908 | 0.37309 | 0.38322 | 0.25577 |
| 9 | 680 | 0.46698 | 0.44533 | 0.24812 |
| 10 | 1178 | 0.33544 | 0.51129 | 0.26289 |
| 11 | 5690 | 0.28409 | 0.29009 | 0.24376 |
| 12 | 9385 | 0.36731 | 0.23495 | 0.23873 |
| 13 | 6878 | 0.31962 | 0.31088 | 0.23691 |
| 14 | 1749 | 0.43068 | 0.47078 | 0.25696 |
| 15 | 1870 | 0.29306 | 0.46181 | 0.26494 |
| 16 | 6328 | 0.19137 | 0.29002 | 0.21465 |
| 17 | 7599 | 0.17418 | 0.1739 | 0.22093 |
| 18 | 7072 | 0.36776 | 0.18662 | 0.20821 |
| 19 | 2609 | 0.37377 | 0.42663 | 0.24206 |
| 20 | 1806 | 0.40342 | 0.57026 | 0.32007 |
| 21 | 4247 | 0.35488 | 0.34512 | 0.19181 |
| 22 | 3558 | 0.21403 | 0.30996 | 0.20695 |
| 23 | 4103 | 0.36559 | 0.19154 | 0.17851 |
| 24 | 2270 | 0.25744 | 0.48077 | 0.26616 |
| 25 | 480 | 0.38505 | 1.0035 | 0.73284 |
| 26 | 930 | 0.15033 | 0.26661 | 0.34611 |
| 27 | 971 | 0.11836 | 0.19105 | 0.18709 |
| 28 | 975 | 0.11907 | 0.25901 | 0.30046 |
| 29 | 549 | 0.41024 | 0.95027 | 0.72705 |
| 30 | 1478 | 0.56625 | 0.3835 | 0.34664 |
| 31 | 3216 | 0.16373 | 0.19856 | 0.19257 |
| 32 | 2826 | 0.20149 | 0.14689 | 0.19485 |
| 33 | 3285 | 0.12942 | 0.30274 | 0.20848 |
| 34 | 1965 | 0.38874 | 0.45564 | 0.38875 |
| 35 | 1934 | 0.61707 | 0.35339 | 0.25067 |
| 36 | 5875 | 0.55413 | 0.29556 | 0.19246 |
| 37 | 5992 | 0.2137 | 0.12029 | 0.21502 |
| 38 | 6589 | 0.2254 | 0.21047 | 0.21193 |
| 39 | 2763 | 0.39851 | 0.41673 | 0.27272 |
| 40 | 1512 | 0.50654 | 0.36893 | 0.25051 |
| 41 | 6476 | 0.28308 | 0.21635 | 0.22668 |
| 42 | 9212 | 0.40577 | 0.18283 | 0.2252 |
| 43 | 7521 | 0.22936 | 0.29881 | 0.23864 |
| 44 | 2392 | 0.40816 | 0.42881 | 0.2751 |
| 45 | 611 | 0.56917 | 0.48084 | 0.26191 |
| 46 | 4379 | 0.42672 | 0.42447 | 0.25285 |
| 47 | 7749 | 0.29309 | 0.4378 | 0.24631 |
| 48 | 5470 | 0.67537 | 0.37528 | 0.25187 |
| 49 | 1210 | 0.42778 | 0.53938 | 0.27266 |
| 50 | 127 | 0.92194 | 0.45501 | 0.24629 |
| 51 | 2235 | 0.71785 | 0.33424 | 0.25781 |
| 52 | 4379 | 0.41055 | 0.38155 | 0.25735 |
| 53 | 2757 | 0.44268 | 0.40149 | 0.26646 |
| 54 | 327 | 0.52856 | 0.51306 | 0.28205 |
| 56 | 457 | 0.60967 | 0.429 | 0.26482 |
| 57 | 1507 | 0.70752 | 0.67612 | 0.25844 |
| 58 | 786 | 0.53669 | 0.50635 | 0.27127 |
| 59 | 39 | 0.43205 | 0.65796 | 0.25585 |
-----------------------------------------------
Saving refined experiments to refined_lev0.expt
Saving refined reflections to refined_lev0.refl
Outlier rejection has cleaned up the positional residuals so now the greatest deviation is within 0.4 mm of the predicted position. The angular extreme is now just over 0.4 degrees. Coverage of the outer and central panels (where reflections are in the backstop shadow or thrown away for being too close to the spindle) is still a little low. Notably, panel 55 (a corner panel) is completely missing. If we had more datasets recorded at the same detector distance (and more time to process them) we could combine them in a multi-crystal joint refinement job to increase the coverage of panels further. However, for the purposes of this tutorial we will see what we can get with this single dataset.
Before moving on to the multi-panel refinement job we will take a look at the refined reflections file:
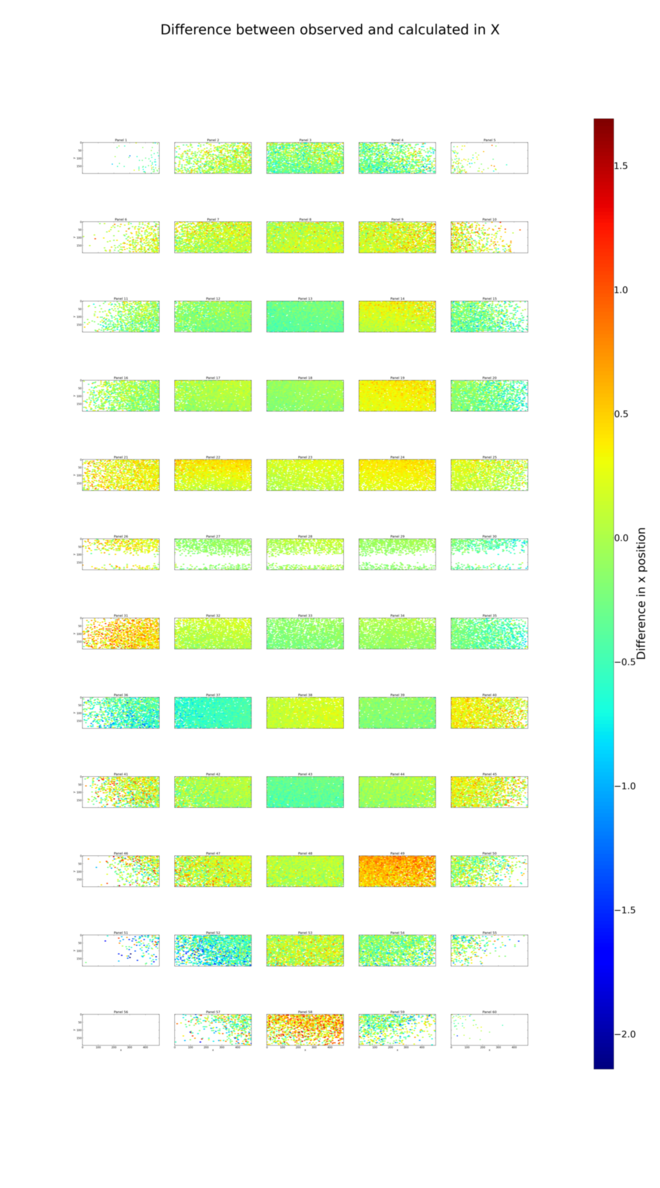
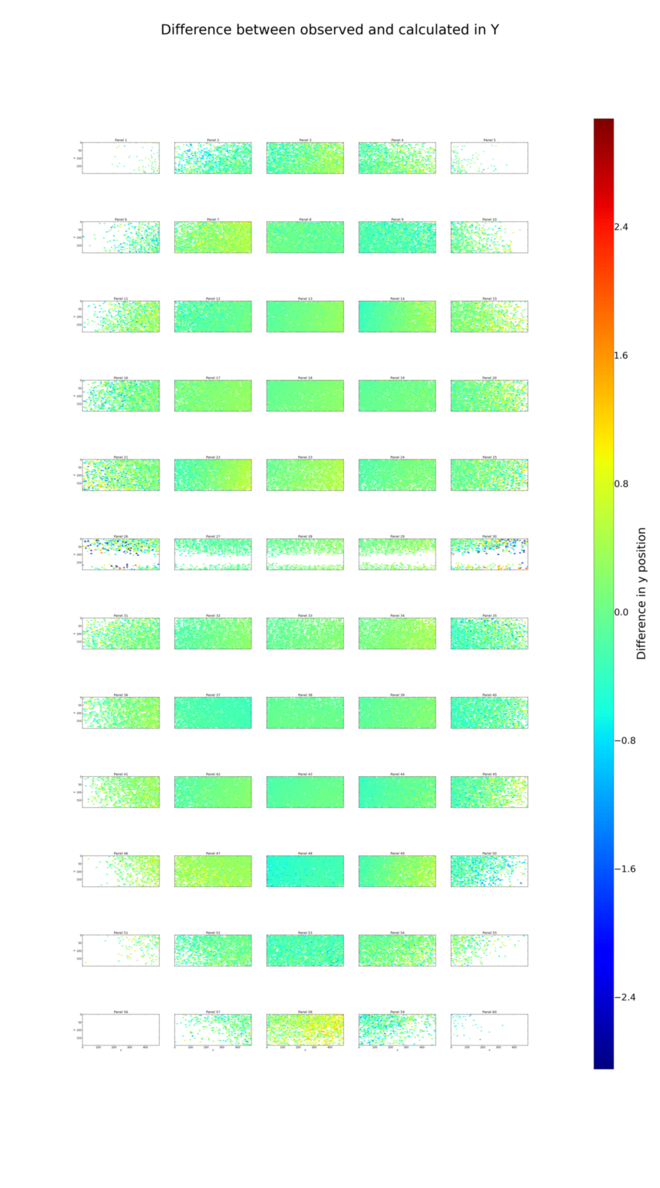
dials.analyse_output refined_lev0.refl grid_size=5,12
Here we had to tell dials.analyse_output about the arrangement of
the panels, as it does not use the refined_lev0.expt file so cannot
figure this out itself.
Here are the positional residual plots for X and Y, analysis/centroid/centroid_diff_x.png
and analysis/centroid/centroid_diff_y.png. The multi-panel versions
of these plots are not as compact as the single tile version presented in the
Processing in Detail. However, careful comparison of the plots is enough to
show that the same pattern of shifts is present.
Multi-tile refinement¶
Now we repeat refinement, but we allow the panels to move independently. In
DIALS multi-panel detectors are represented by a hierarchical model. The highest
level hieararchy_level=0 means to treat the whole detector unit as a
rigid block. Some detectors, notably the CS-PAD used at LCLS beamlines, have a
real hierarchy of a few levels deep. The Pilatus P6M has a very simple hierarchy,
with a single lower level, hieararchy_level=1, in which every panel is
treated separately. We now start from the previous refinement run
specifying this hierarchy level:
dials.refine indexed.refl refined_lev0.expt outlier.algorithm=tukey \
use_all_reflections=true output.reflections=refined_lev1.refl \
close_to_spindle_cutoff=0.01 bin_size_fraction=0 hierarchy_level=1 \
output.experiments=refined_lev1.expt
You may have noticed that apart from hierarchy_level=1 there was an
additional parameter added to this command compared to the previous refinement run,
namely bin_size_fraction=0. This sets the RMSD target for refinement to
zero, so that refinement will never terminate because the RMSDs are ‘good enough’,
only if they converge so that their rate of decrease on subsequent steps falls
to zero. This is necessary because the extra freedom allowed by parameterising
each panel individually allows the RMSDs to fall lower than the default target.
There are 366 parameters in total for this refinement run. This can be seen
by checking the file dials.refine.debug.log once refinement is underway.
Warning
This job took 17 minutes to run on a Linux desktop with a Core i7 CPU running at 3.07GHz, and uses about 4 GB of RAM.
Refinement is single-process at the moment, unfortunately, so we can’t yet make use of parallelism here to speed the job up. The output is as follows:
The following parameters have been modified:
output {
experiments = refined_lev1.expt
reflections = refined_lev1.refl
}
refinement {
parameterisation {
detector {
hierarchy_level = 1
}
}
target {
bin_size_fraction = 0
}
reflections {
use_all_reflections = true
close_to_spindle_cutoff = 0.01
outlier {
algorithm = null *tukey
}
}
}
input {
experiments = refined.expt
reflections = indexed.refl
}
Configuring refiner
Summary statistics for observations matched to predictions:
------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
------------------------------------------------------------------------
| Xc - Xo (mm) | -2.496 | -0.04178 | 0.0004518 | 0.04222 | 2.133 |
| Yc - Yo (mm) | -1.903 | -0.03577 | 0.0006705 | 0.03588 | 1.656 |
| Phic - Phio (deg) | -5.576 | -0.06467 | -0.0007391 | 0.06414 | 3.292 |
| X weights | 80.55 | 127.9 | 131.9 | 134 | 135.2 |
| Y weights | 77.02 | 127.7 | 131.9 | 134 | 135.2 |
| Phi weights | 25.97 | 32.57 | 32.65 | 32.65 | 32.65 |
------------------------------------------------------------------------
6433 reflections have been rejected as outliers
Summary statistics for observations matched to predictions:
--------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
--------------------------------------------------------------------------
| Xc - Xo (mm) | -0.2916 | -0.04138 | 0.0001204 | 0.041 | 0.3683 |
| Yc - Yo (mm) | -0.4838 | -0.03481 | 0.0008164 | 0.03518 | 0.4917 |
| Phic - Phio (deg) | -0.6969 | -0.06416 | -0.0008647 | 0.06351 | 0.7644 |
| X weights | 80.55 | 128 | 132 | 134 | 135.2 |
| Y weights | 77.02 | 127.8 | 131.9 | 134.1 | 135.2 |
| Phi weights | 26.58 | 32.57 | 32.65 | 32.65 | 32.65 |
--------------------------------------------------------------------------
Performing refinement...
Refinement steps:
--------------------------------------------------
| Step | Nref | RMSD_X | RMSD_Y | RMSD_Phi |
| | | (mm) | (mm) | (deg) |
--------------------------------------------------
| 0 | 186145 | 0.063617 | 0.058127 | 0.085801 |
| 1 | 186145 | 0.056976 | 0.05538 | 0.085719 |
| 2 | 186145 | 0.049808 | 0.052619 | 0.085597 |
| 3 | 186145 | 0.04634 | 0.051408 | 0.085475 |
| 4 | 186145 | 0.045568 | 0.051142 | 0.085391 |
| 5 | 186145 | 0.04538 | 0.051014 | 0.085337 |
| 6 | 186145 | 0.045228 | 0.050729 | 0.085285 |
| 7 | 186145 | 0.045054 | 0.050076 | 0.085215 |
| 8 | 186145 | 0.044868 | 0.049034 | 0.085115 |
| 9 | 186145 | 0.044746 | 0.048167 | 0.085025 |
| 10 | 186145 | 0.044708 | 0.047833 | 0.08498 |
| 11 | 186145 | 0.044695 | 0.047759 | 0.084969 |
| 12 | 186145 | 0.04468 | 0.047738 | 0.084967 |
| 13 | 186145 | 0.044668 | 0.047726 | 0.084967 |
| 14 | 186145 | 0.044662 | 0.047721 | 0.084967 |
| 15 | 186145 | 0.04466 | 0.047719 | 0.084967 |
--------------------------------------------------
RMSD no longer decreasing
RMSDs by experiment:
-----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
-----------------------------------------------
| 0 | 186145 | 0.25965 | 0.27743 | 0.24276 |
-----------------------------------------------
RMSDs by panel:
------------------------------------------------
| Panel | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
------------------------------------------------
| 0 | 64 | 0.32036 | 0.57819 | 0.22482 |
| 1 | 1361 | 0.34304 | 0.4111 | 0.26504 |
| 2 | 3079 | 0.3561 | 0.36483 | 0.26682 |
| 3 | 1632 | 0.37843 | 0.37838 | 0.25453 |
| 4 | 112 | 0.39065 | 0.39675 | 0.23706 |
| 5 | 394 | 0.30539 | 0.48132 | 0.25442 |
| 6 | 3490 | 0.29682 | 0.35249 | 0.25661 |
| 7 | 6178 | 0.28525 | 0.27486 | 0.25318 |
| 8 | 3909 | 0.31728 | 0.30551 | 0.25565 |
| 9 | 675 | 0.39465 | 0.42802 | 0.24507 |
| 10 | 1175 | 0.30057 | 0.46365 | 0.26215 |
| 11 | 5685 | 0.23485 | 0.27143 | 0.24326 |
| 12 | 9386 | 0.20171 | 0.18887 | 0.23872 |
| 13 | 6880 | 0.23494 | 0.23081 | 0.23689 |
| 14 | 1746 | 0.31911 | 0.40137 | 0.25357 |
| 15 | 1870 | 0.27575 | 0.44911 | 0.26334 |
| 16 | 6321 | 0.18338 | 0.21691 | 0.21382 |
| 17 | 7608 | 0.14673 | 0.13459 | 0.22118 |
| 18 | 7075 | 0.15323 | 0.1772 | 0.20816 |
| 19 | 2608 | 0.27977 | 0.40106 | 0.23663 |
| 20 | 1803 | 0.24748 | 0.54944 | 0.31271 |
| 21 | 4249 | 0.15151 | 0.21436 | 0.18971 |
| 22 | 3560 | 0.11336 | 0.12431 | 0.20676 |
| 23 | 4103 | 0.10087 | 0.16239 | 0.17534 |
| 24 | 2271 | 0.20061 | 0.46907 | 0.24929 |
| 25 | 476 | 0.26774 | 0.94809 | 0.69743 |
| 26 | 927 | 0.10167 | 0.22193 | 0.33398 |
| 27 | 974 | 0.097735 | 0.13559 | 0.1876 |
| 28 | 977 | 0.069491 | 0.1907 | 0.29589 |
| 29 | 549 | 0.19349 | 0.91362 | 0.67233 |
| 30 | 1474 | 0.31601 | 0.36268 | 0.32879 |
| 31 | 3209 | 0.13728 | 0.15038 | 0.18643 |
| 32 | 2829 | 0.10162 | 0.11078 | 0.19453 |
| 33 | 3276 | 0.10363 | 0.17357 | 0.2043 |
| 34 | 1951 | 0.20521 | 0.4038 | 0.36983 |
| 35 | 1931 | 0.37408 | 0.28915 | 0.24379 |
| 36 | 5869 | 0.18862 | 0.14476 | 0.1912 |
| 37 | 5991 | 0.13887 | 0.11716 | 0.21506 |
| 38 | 6591 | 0.15001 | 0.19861 | 0.21179 |
| 39 | 2763 | 0.25238 | 0.36458 | 0.26476 |
| 40 | 1511 | 0.47841 | 0.27181 | 0.24826 |
| 41 | 6461 | 0.27368 | 0.18468 | 0.22587 |
| 42 | 9213 | 0.19848 | 0.15776 | 0.22531 |
| 43 | 7521 | 0.21923 | 0.24706 | 0.2383 |
| 44 | 2390 | 0.32645 | 0.40052 | 0.27085 |
| 45 | 611 | 0.55212 | 0.29673 | 0.26008 |
| 46 | 4378 | 0.38894 | 0.24988 | 0.25198 |
| 47 | 7747 | 0.2824 | 0.2289 | 0.24614 |
| 48 | 5470 | 0.29905 | 0.31716 | 0.25135 |
| 49 | 1210 | 0.41311 | 0.47572 | 0.2691 |
| 50 | 127 | 0.69191 | 0.39228 | 0.2447 |
| 51 | 2231 | 0.47797 | 0.32129 | 0.25717 |
| 52 | 4378 | 0.38899 | 0.31331 | 0.25676 |
| 53 | 2758 | 0.39246 | 0.40955 | 0.26575 |
| 54 | 327 | 0.48186 | 0.52671 | 0.2798 |
| 56 | 458 | 0.57234 | 0.42803 | 0.2643 |
| 57 | 1507 | 0.494 | 0.41177 | 0.25741 |
| 58 | 786 | 0.48028 | 0.48906 | 0.26898 |
| 59 | 40 | 0.39499 | 0.38962 | 0.2466 |
------------------------------------------------
Saving refined experiments to refined_lev1.expt
Saving refined reflections to refined_lev1.refl
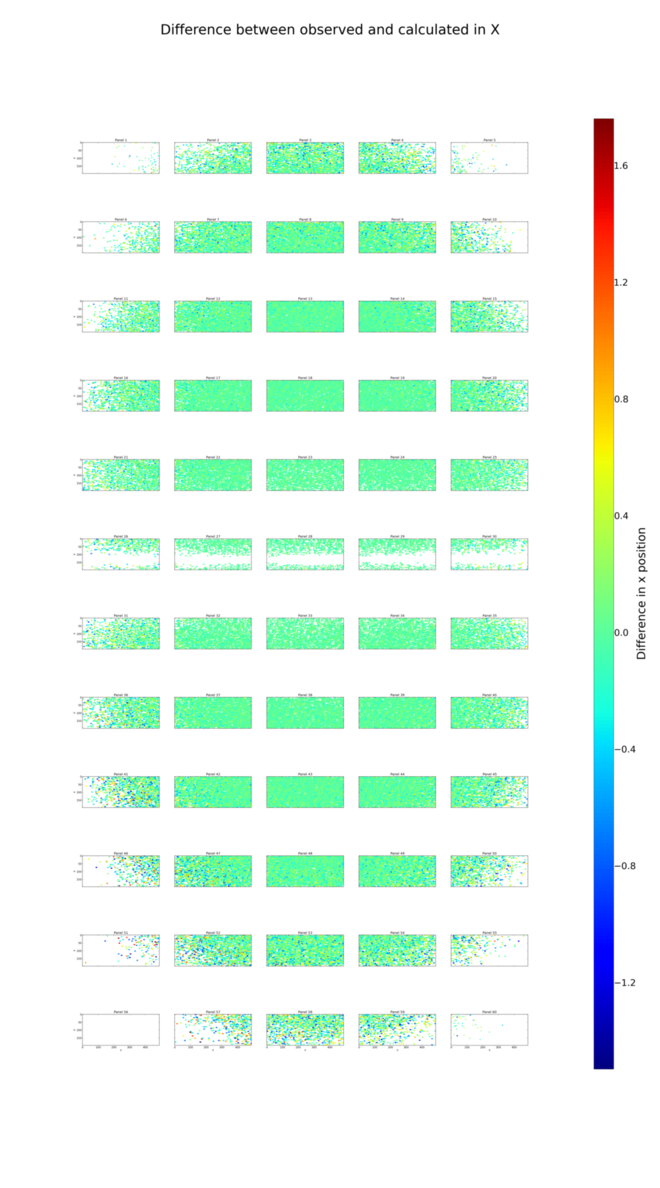
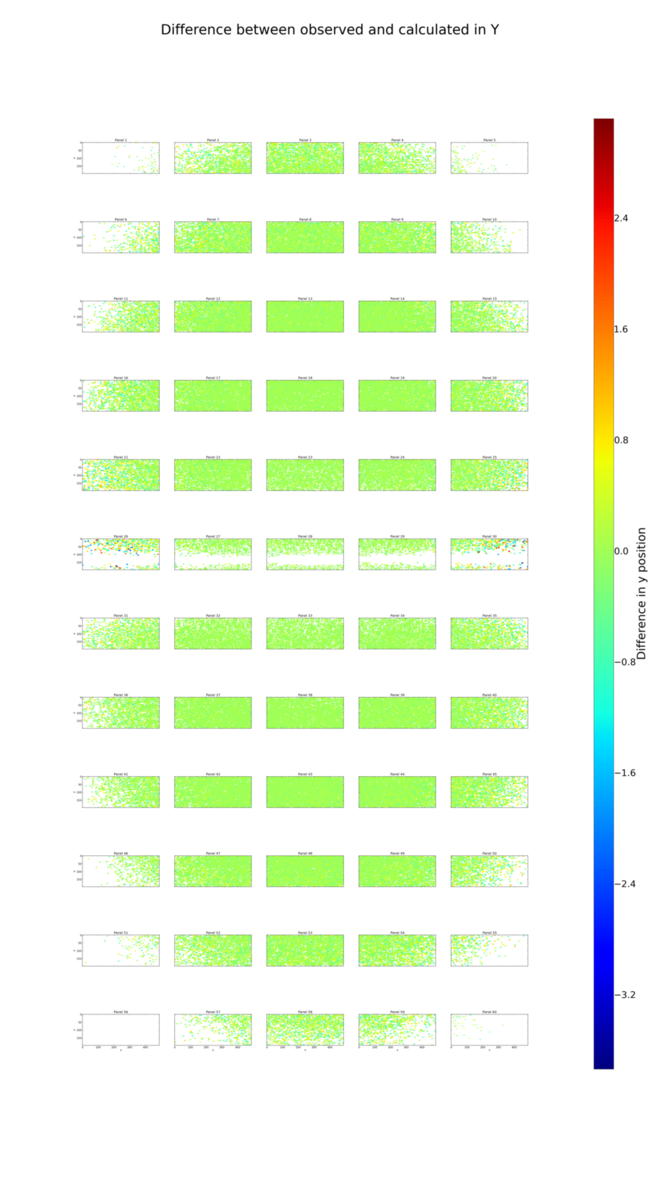
Following refinement, we repeat the analysis of positional residuals:
mv analysis analysis_lev0
dials.analyse_output refined.refl grid_size=5,12
mv analysis analysis_lev1
The positional residual plots for X and Y,
analysis_lev1/centroid/centroid_diff_x.png
and analysis_lev1/centroid/centroid_diff_y.png make it clear that
despite poor coverage on some panels, the systematic shifts have been cleaned
up by the refinement job.
Applying the corrected metrology¶
Although we have corrected for small shifts and rotations apparently present from spot positions recorded in one dataset, what we would really like to do is apply these corrections to a different dataset. Short of rewriting the dxtbx Format object for the detector to incorporate the corrections, we could try to ‘copy and paste’ the detector from one dataset to the other.
We will choose the standard tutorial data to try this, from the
Processing in Detail. First we have to process that data using the
multi-panel version of the Pilatus P6M detector model. Assuming the environment
variable P6M_60_PANEL=1 is set in this terminal we just need to repeat
the commands from the tutorial:
mkdir tutorial_data
cd !$
dials.import /path/to/th_8_2*cbf
dials.find_spots imported.expt nproc=4
dials.index imported.expt strong.refl space_group="P4"
dials.refine indexed.expt indexed.refl outlier.algorithm=tukey use_all_reflections=true bin_size_fraction=0.0
Note these are the overall RMSDs (comparable to the results from the Processing in Detail, as we’d expect):
RMSDs by experiment:
----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
----------------------------------------------
| 0 | 53916 | 0.26341 | 0.21858 | 0.106 |
----------------------------------------------
Now we do the scan-varying refinement and integrate:
dials.refine refined.expt indexed.refl outlier.algorithm=tukey use_all_reflections=true bin_size_fraction=0.0 scan_varying=true output.experiments=sv_refined.expt
dials.integrate sv_refined.expt indexed.refl outlier.algorithm=null nproc=4
dials.export integrated.refl sv_refined.expt mtz.hklout=integrated.mtz
dials.analyse_output integrated.refl grid_size=5,12
From the end of dials.integrate.log:
Summary of integration results binned by resolution
----------------------------------------------------------------------------------------------------------
d min | d max | # full | # part | # over | # ice | # sum | # prf | <Ibg> | <I/sigI> | <I/sigI> | <CC prf>
| | | | | | | | | (sum) | (prf) |
----------------------------------------------------------------------------------------------------------
1.17 | 1.19 | 324 | 3 | 0 | 0 | 327 | 260 | 0.04 | 0.37 | 0.54 | 0.11
1.19 | 1.21 | 1093 | 7 | 0 | 0 | 1100 | 984 | 0.04 | 0.43 | 0.54 | 0.09
1.21 | 1.23 | 2317 | 14 | 0 | 0 | 2331 | 2138 | 0.05 | 0.48 | 0.57 | 0.10
1.23 | 1.26 | 3776 | 27 | 0 | 0 | 3803 | 3581 | 0.05 | 0.53 | 0.66 | 0.12
1.26 | 1.28 | 5416 | 34 | 0 | 0 | 5450 | 5100 | 0.05 | 0.59 | 0.74 | 0.14
1.28 | 1.31 | 7235 | 54 | 0 | 0 | 7289 | 6806 | 0.06 | 0.64 | 0.81 | 0.16
1.31 | 1.35 | 9504 | 63 | 0 | 0 | 9567 | 8952 | 0.06 | 0.77 | 0.95 | 0.19
1.35 | 1.38 | 12495 | 87 | 0 | 0 | 12582 | 11826 | 0.07 | 0.90 | 1.11 | 0.21
1.38 | 1.42 | 16967 | 111 | 0 | 0 | 17078 | 16125 | 0.08 | 0.98 | 1.20 | 0.23
1.42 | 1.47 | 20187 | 153 | 0 | 0 | 20340 | 19437 | 0.08 | 1.20 | 1.43 | 0.27
1.47 | 1.52 | 23623 | 258 | 0 | 0 | 23881 | 22761 | 0.09 | 1.47 | 1.71 | 0.30
1.52 | 1.58 | 24126 | 271 | 0 | 0 | 24397 | 23289 | 0.10 | 1.75 | 2.03 | 0.35
1.58 | 1.66 | 25506 | 281 | 0 | 0 | 25787 | 24863 | 0.11 | 2.17 | 2.46 | 0.41
1.66 | 1.74 | 24269 | 241 | 0 | 0 | 24510 | 23389 | 0.12 | 2.71 | 3.01 | 0.46
1.74 | 1.85 | 24780 | 238 | 0 | 0 | 25018 | 24019 | 0.15 | 3.50 | 3.82 | 0.52
1.85 | 2.00 | 25710 | 278 | 0 | 0 | 25988 | 25059 | 0.19 | 4.87 | 5.20 | 0.59
2.00 | 2.20 | 24757 | 274 | 0 | 0 | 25031 | 24000 | 0.26 | 6.58 | 6.94 | 0.66
2.20 | 2.51 | 25673 | 252 | 0 | 0 | 25925 | 25119 | 0.29 | 8.84 | 9.15 | 0.69
2.51 | 3.17 | 25214 | 297 | 0 | 0 | 25511 | 24607 | 0.37 | 12.78 | 13.01 | 0.73
3.17 | 151.26 | 25766 | 303 | 0 | 0 | 26069 | 25226 | 0.45 | 25.48 | 25.36 | 0.74
----------------------------------------------------------------------------------------------------------
Summary of integration results for the whole dataset
----------------------------------------------
Number fully recorded | 338720
Number partially recorded | 4747
Number with overloaded pixels | 0
Number in powder rings | 0
Number processed with summation | 331984
Number processed with profile fitting | 317541
<Ibg> | 0.18
<I/sigI> (summation) | 5.63
<I/sigI> (profile fitting) | 5.92
<CC prf> | 0.43
----------------------------------------------
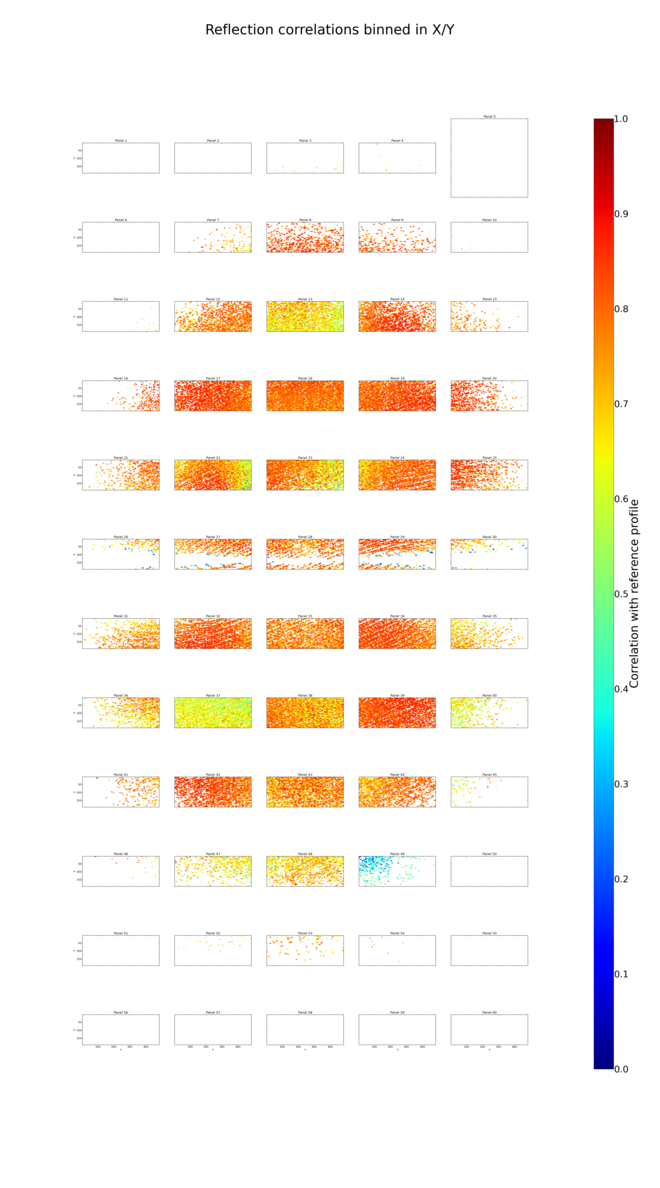
The plot of correlations between strong (reference) reflections and the profiles used to fit them shows up the systematic effects of uncorrected tile shifts and misorientations:
Now, how can we apply the metrology? Here we will use dials.combine_experiments
with the reference_from_experiment.detector option to overwrite the detector
model from our indexed.expt. We don’t really want the combined experiments
file, only this side-effect, so we immediately split it again:
dials.combine_experiments experiments=../refined_lev1.expt experiments=refined.expt reflections=../refined_lev1.refl reflections=indexed.refl reference_from_experiment.detector=0
dials.split_experiments combined.expt combined.refl
This results in a few files, of which experiments_1.expt is interesting.
It contains the updated detector - but beware the detector distance is now
completely wrong! It is at about 400 mm rather than 265 mm. We could correct that
by editing experiments_1.expt directly, but actually there is no need.
dials.refine is extremely forgiving of bad starting geometry, though
we should remember to fix the beam and crystal models:
dials.refine experiments_1.expt indexed.refl output.experiments=corrected_refined.expt beam.fix=all crystal.fix=all
A snippet from the log file shows that the detector distance offset was largely corrected in a single step:
Refinement steps:
------------------------------------------------
| Step | Nref | RMSD_X | RMSD_Y | RMSD_Phi |
| | | (mm) | (mm) | (deg) |
------------------------------------------------
| 0 | 4049 | 40.622 | 40.688 | 0.019202 |
| 1 | 4049 | 0.28874 | 0.30251 | 0.019202 |
| 2 | 4049 | 0.045937 | 0.045205 | 0.019202 |
------------------------------------------------
Now we’ll let the crystal and beam refine along with the new detector to RMSD convergence:
dials.refine corrected_refined.expt indexed.refl outlier.algorithm=tukey use_all_reflections=true bin_size_fraction=0.0 output.experiments=corrected_refined.expt
Here is the output:
The following parameters have been modified:
output {
experiments = corrected_refined.expt
}
refinement {
target {
bin_size_fraction = 0.0
}
reflections {
use_all_reflections = true
outlier {
algorithm = null *tukey
}
}
}
input {
experiments = corrected_refined.expt
reflections = indexed.refl
}
Configuring refiner
Summary statistics for observations matched to predictions:
-------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
-------------------------------------------------------------------------
| Xc - Xo (mm) | -0.4701 | -0.02988 | -0.01025 | 0.02159 | 0.6902 |
| Yc - Yo (mm) | -0.7267 | -0.02846 | -0.005739 | 0.02339 | 1.247 |
| Phic - Phio (deg) | -1.409 | -0.01035 | 5.562e-05 | 0.01104 | 0.4029 |
| X weights | 113.5 | 133.7 | 134.5 | 135 | 135.2 |
| Y weights | 109.2 | 133.4 | 134.5 | 135 | 135.2 |
| Phi weights | 155.5 | 175.8 | 177 | 177.5 | 177.8 |
-------------------------------------------------------------------------
2382 reflections have been rejected as outliers
Summary statistics for observations matched to predictions:
--------------------------------------------------------------------------
| | Min | Q1 | Med | Q3 | Max |
--------------------------------------------------------------------------
| Xc - Xo (mm) | -0.1148 | -0.02997 | -0.01092 | 0.01966 | 0.331 |
| Yc - Yo (mm) | -0.1397 | -0.02791 | -0.005742 | 0.02263 | 0.2531 |
| Phic - Phio (deg) | -0.1881 | -0.009998 | 0.0001419 | 0.0109 | 0.1176 |
| X weights | 113.5 | 133.7 | 134.6 | 135 | 135.2 |
| Y weights | 109.9 | 133.5 | 134.5 | 135 | 135.2 |
| Phi weights | 158.2 | 175.9 | 177 | 177.5 | 177.8 |
--------------------------------------------------------------------------
Performing refinement...
Refinement steps:
-------------------------------------------------
| Step | Nref | RMSD_X | RMSD_Y | RMSD_Phi |
| | | (mm) | (mm) | (deg) |
-------------------------------------------------
| 0 | 53738 | 0.04524 | 0.04251 | 0.015875 |
| 1 | 53738 | 0.043495 | 0.041514 | 0.015395 |
| 2 | 53738 | 0.042657 | 0.040762 | 0.01535 |
| 3 | 53738 | 0.040455 | 0.038826 | 0.015287 |
| 4 | 53738 | 0.0356 | 0.034575 | 0.015251 |
| 5 | 53738 | 0.028146 | 0.027998 | 0.015246 |
| 6 | 53738 | 0.021919 | 0.022913 | 0.015245 |
| 7 | 53738 | 0.019867 | 0.021931 | 0.015256 |
| 8 | 53738 | 0.019579 | 0.02202 | 0.015267 |
| 9 | 53738 | 0.019553 | 0.02204 | 0.015269 |
| 10 | 53738 | 0.019552 | 0.022041 | 0.015269 |
-------------------------------------------------
RMSD no longer decreasing
RMSDs by experiment:
----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
----------------------------------------------
| 0 | 53738 | 0.11368 | 0.12815 | 0.10179 |
----------------------------------------------
RMSDs by panel:
-------------------------------------------------
| Panel | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
-------------------------------------------------
| 2 | 18 | 0.18212 | 0.28586 | 0.12145 |
| 6 | 125 | 0.15295 | 0.24972 | 0.12934 |
| 7 | 609 | 0.1286 | 0.21324 | 0.12291 |
| 8 | 265 | 0.1243 | 0.21443 | 0.10878 |
| 11 | 1189 | 0.14339 | 0.19084 | 0.10244 |
| 12 | 2389 | 0.097393 | 0.16185 | 0.1081 |
| 13 | 1550 | 0.084417 | 0.14168 | 0.098364 |
| 14 | 148 | 0.14653 | 0.17253 | 0.11116 |
| 15 | 254 | 0.21387 | 0.15463 | 0.10746 |
| 16 | 2728 | 0.11345 | 0.15427 | 0.081722 |
| 17 | 4265 | 0.060489 | 0.1104 | 0.096919 |
| 18 | 2952 | 0.07167 | 0.092076 | 0.093342 |
| 19 | 565 | 0.1626 | 0.11391 | 0.11975 |
| 20 | 534 | 0.18454 | 0.16728 | 0.16016 |
| 21 | 2772 | 0.092555 | 0.12647 | 0.076534 |
| 22 | 2873 | 0.047736 | 0.10398 | 0.090391 |
| 23 | 2686 | 0.063075 | 0.074814 | 0.075038 |
| 24 | 729 | 0.16869 | 0.11128 | 0.14306 |
| 25 | 119 | 0.81495 | 0.29087 | 0.44711 |
| 26 | 406 | 0.080441 | 0.12121 | 0.098888 |
| 27 | 425 | 0.052654 | 0.15365 | 0.0754 |
| 28 | 364 | 0.06931 | 0.066238 | 0.073655 |
| 29 | 100 | 0.85448 | 0.22713 | 0.24889 |
| 30 | 579 | 0.21735 | 0.11564 | 0.18439 |
| 31 | 1994 | 0.061756 | 0.09509 | 0.068866 |
| 32 | 1924 | 0.091952 | 0.11146 | 0.083513 |
| 33 | 2166 | 0.088252 | 0.10009 | 0.085247 |
| 34 | 519 | 0.13737 | 0.16646 | 0.1872 |
| 35 | 634 | 0.16379 | 0.093519 | 0.12164 |
| 36 | 2835 | 0.067371 | 0.080697 | 0.081958 |
| 37 | 3737 | 0.094318 | 0.086918 | 0.10408 |
| 38 | 2806 | 0.098988 | 0.13001 | 0.082836 |
| 39 | 417 | 0.17099 | 0.18814 | 0.12774 |
| 40 | 227 | 0.20834 | 0.14785 | 0.10324 |
| 41 | 1817 | 0.082715 | 0.1174 | 0.10219 |
| 42 | 2385 | 0.1096 | 0.12543 | 0.10965 |
| 43 | 1589 | 0.12167 | 0.17861 | 0.099929 |
| 44 | 88 | 0.21088 | 0.2111 | 0.10221 |
| 45 | 28 | 0.19222 | 0.16491 | 0.16288 |
| 46 | 472 | 0.094714 | 0.15589 | 0.1232 |
| 47 | 1047 | 0.11775 | 0.17961 | 0.12234 |
| 48 | 307 | 0.12735 | 0.22443 | 0.11915 |
| 51 | 22 | 0.11972 | 0.18238 | 0.11037 |
| 52 | 80 | 0.12845 | 0.24552 | 0.1396 |
-------------------------------------------------
Saving refined experiments to corrected_refined.expt
As a reminder, before metrology correction we had these refined RMSDs from scan- static refinement:
RMSDs by experiment:
----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
----------------------------------------------
| 0 | 53916 | 0.26341 | 0.21858 | 0.106 |
----------------------------------------------
After correction they are as follows:
RMSDs by experiment:
----------------------------------------------
| Exp | Nref | RMSD_X | RMSD_Y | RMSD_Z |
| | | (px) | (px) | (images) |
----------------------------------------------
| 0 | 53738 | 0.11368 | 0.12815 | 0.10179 |
----------------------------------------------
Let’s now do scan-varying refinement then integrate the dataset with corrected metrology:
dials.refine corrected_refined.expt indexed.refl outlier.algorithm=tukey use_all_reflections=true bin_size_fraction=0.0 scan_varying=true output.experiments=corrected_sv_refined.expt
dials.integrate corrected_sv_refined.expt indexed.refl outlier.algorithm=null nproc=4 output.reflections=corrected_integrated.refl
dials.export corrected_integrated.refl corrected_sv_refined.expt mtz.hklout=corrected_integrated.mtz
dials.analyse_output corrected_integrated.refl grid_size=5,12
From the integration log:
Summary of integration results binned by resolution
----------------------------------------------------------------------------------------------------------
d min | d max | # full | # part | # over | # ice | # sum | # prf | <Ibg> | <I/sigI> | <I/sigI> | <CC prf>
| | | | | | | | | (sum) | (prf) |
----------------------------------------------------------------------------------------------------------
1.17 | 1.19 | 340 | 3 | 0 | 0 | 343 | 278 | 0.04 | 0.28 | 0.41 | 0.06
1.19 | 1.21 | 1125 | 6 | 0 | 0 | 1131 | 1011 | 0.05 | 0.37 | 0.42 | 0.06
1.21 | 1.23 | 2346 | 13 | 0 | 0 | 2359 | 2176 | 0.05 | 0.47 | 0.49 | 0.07
1.23 | 1.26 | 3776 | 24 | 0 | 0 | 3800 | 3565 | 0.05 | 0.50 | 0.61 | 0.10
1.26 | 1.29 | 5448 | 37 | 0 | 0 | 5485 | 5123 | 0.05 | 0.58 | 0.71 | 0.12
1.29 | 1.32 | 7241 | 52 | 0 | 0 | 7293 | 6804 | 0.06 | 0.66 | 0.79 | 0.15
1.32 | 1.35 | 9522 | 71 | 0 | 0 | 9593 | 8986 | 0.06 | 0.77 | 0.94 | 0.19
1.35 | 1.38 | 12528 | 84 | 0 | 0 | 12612 | 11851 | 0.07 | 0.92 | 1.11 | 0.22
1.38 | 1.43 | 16954 | 113 | 0 | 0 | 17067 | 16130 | 0.08 | 1.01 | 1.22 | 0.24
1.43 | 1.47 | 20247 | 154 | 0 | 0 | 20401 | 19499 | 0.08 | 1.22 | 1.45 | 0.29
1.47 | 1.52 | 23553 | 267 | 0 | 0 | 23820 | 22740 | 0.09 | 1.50 | 1.74 | 0.33
1.52 | 1.58 | 24022 | 267 | 0 | 0 | 24289 | 23215 | 0.10 | 1.78 | 2.06 | 0.38
1.58 | 1.66 | 25479 | 286 | 0 | 0 | 25765 | 24860 | 0.11 | 2.19 | 2.49 | 0.44
1.66 | 1.74 | 24305 | 241 | 0 | 0 | 24546 | 23395 | 0.12 | 2.74 | 3.05 | 0.51
1.74 | 1.85 | 24742 | 236 | 0 | 0 | 24978 | 23965 | 0.15 | 3.54 | 3.87 | 0.58
1.85 | 2.00 | 25657 | 281 | 0 | 0 | 25938 | 24999 | 0.19 | 4.90 | 5.24 | 0.65
2.00 | 2.20 | 24755 | 268 | 0 | 0 | 25023 | 24005 | 0.26 | 6.61 | 6.97 | 0.72
2.20 | 2.52 | 25596 | 250 | 0 | 0 | 25846 | 25058 | 0.29 | 8.88 | 9.20 | 0.76
2.52 | 3.17 | 25140 | 298 | 0 | 0 | 25438 | 24540 | 0.37 | 12.81 | 13.04 | 0.78
3.17 | 151.25 | 25720 | 303 | 0 | 0 | 26023 | 25193 | 0.44 | 25.52 | 25.39 | 0.79
----------------------------------------------------------------------------------------------------------
Summary of integration results for the whole dataset
----------------------------------------------
Number fully recorded | 338446
Number partially recorded | 4836
Number with overloaded pixels | 0
Number in powder rings | 0
Number processed with summation | 331750
Number processed with profile fitting | 317393
<Ibg> | 0.18
<I/sigI> (summation) | 5.65
<I/sigI> (profile fitting) | 5.94
<CC prf> | 0.47
----------------------------------------------
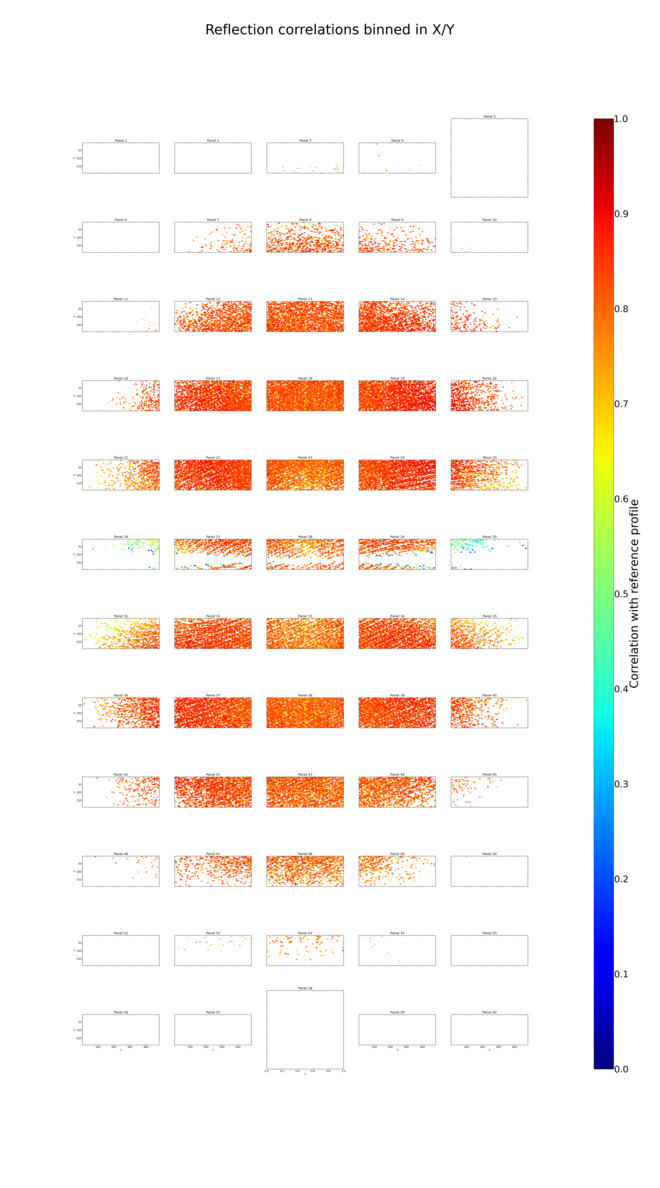
We now have a nicer plot of correlations between profiles and the strong reflections(notwithstanding a few plotting artefacts):
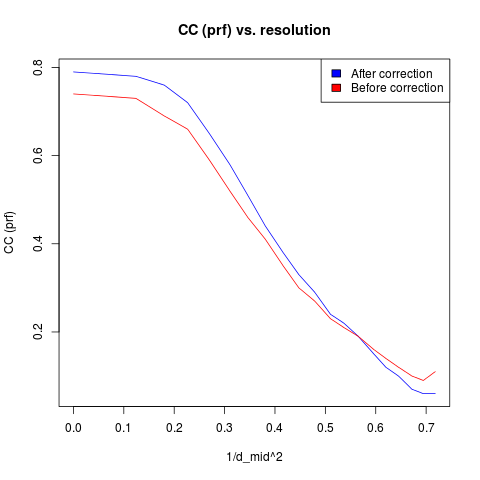
By comparison with the previous results we can see that correcting the panel shifts and mis-orientations has improved the overall profile fitting mean \(\frac{I}{\sigma_I}\) and correlation coefficients. However, comparing the tables of results binned by resolution we see that the improvements are limited to the low resolution, and the results are worse at high resolution. For example, we can make a quick plot of the profile fitting correlation coefficients.
From this we see the crossover point at about \(\frac{1}{d_\textrm{mid}^2} = 0.55\) or a resolution of about 1.35 Angstroms.
So, finally, we conclude that although we have the means to do metrology analysis and corrections of modular detectors, we have learned that it is critical to use data that gives coverage right out to the corners of the detector so that we can apply the corrected metrology to other datasets effectively. With dials.refine this could be achieved by joint refinement of data from multiple crystals, but it will take a significant time to run and use a lot of memory. It is best to do this right, and exactly once!
What to do next¶
Panel coverage was rather low in the corners. We could try combining more metrology datasets together to improve coverage and make an effort at factoring out crystal-dependent effects.
Once we are satisfied with the corrected detector model we can write the changes directly back into the dxtbx Format object, so that they are available immediately for any dataset, without having to perform the ‘copy-and-paste’ operation presented here.
Acknowledgements¶
Dave Hall (Diamond Light Source) for collecting the data.